
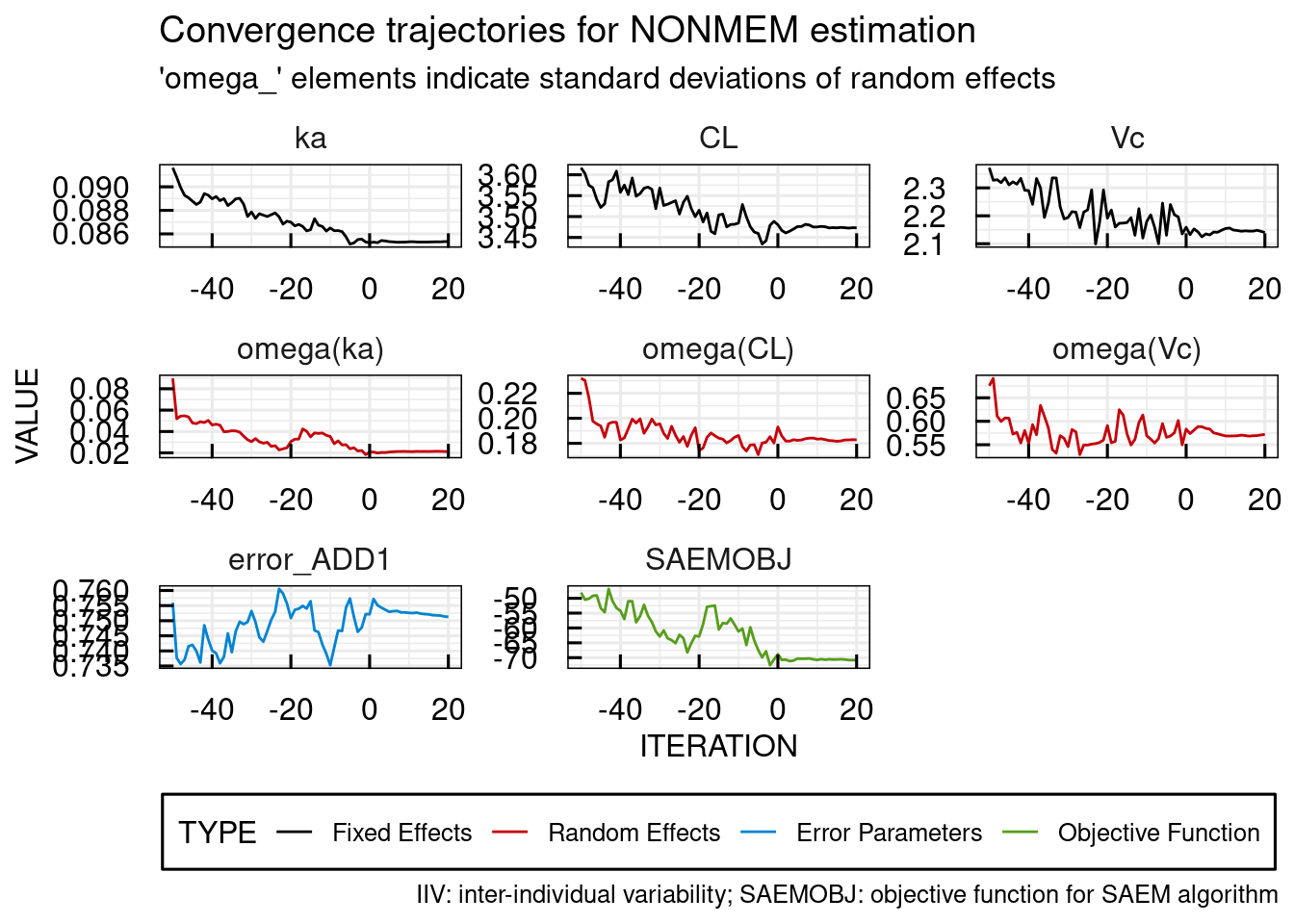
ONEILTable: It is an example data in table format for 2-drug combinations with 4 replicates. SynergyFinderPlus provides 3 example data: tab-delimited TXT file, with extension “.txt”.comma-delimited CSV file, with extension “.csv”.SynergyFinder accepts following file formats: The drug concentrations should be located at the top and left side of the matrix, where concentrations located at the left side correspond to Drug1 and concentrations located at the top correspond to the Drug2.Please use table format for combinations with more than 3 drugs. Matrix format only works on 2 drug combination data.Two dose-response matrices are provided as an example. The three rows below should precede each dose-response matrix:įig.2 An input file in Matrix format. In the Matrix format, the dose-response matrix is represented in a matrix with drug concentrations shown along the left (for Drug1) and top (for Drug2) edges of the matrix (Fig.2). The missing value will be automatically imputed by mice R package.
SynergyFinder allows for missing values in the dose-response matrix.The data should however, contain at least three concentrations for each drug, so that sensible synergy scores can be calculated. There is no restriction on the number of drug combinations for the input file.The duplicated concentration combinations in one block (with the same “block_id”) will be treated as replicates.The cell line names will be used for “data annotation”. Used if the concentration units are not identical across the drugs in test one block. Used for higher-order drug combination data point. Used for higher-order drug combination data point.Ĭoncentration of n th tested drug. For example, “drug3” for the third tested drug. Unit of concentration for the second drug. Used if the concentration units are not identical across the drugs tested in one block. Unit of concentration for the first drug. This column could be replaced by multiple separated columns for each tested drugs (see table below), while different unit was used for measuring the concentrations. Identifier for the drug combination blocks.Ĭell response to the drug treatment (%inhibition or %viability). 'Alternative Column names' are accepted): Required Columns The input table must contain the following columns (The column naming style used in the old SynergyFinder or DrugComb, i.e. In the Table format, the dose-response data is represented as a long table where each row represent one observation in the dose-response matrix (Fig.1). SynergyFinder accepts two formats of input: Table and Matrix.


 0 kommentar(er)
0 kommentar(er)
